Help Tutorial
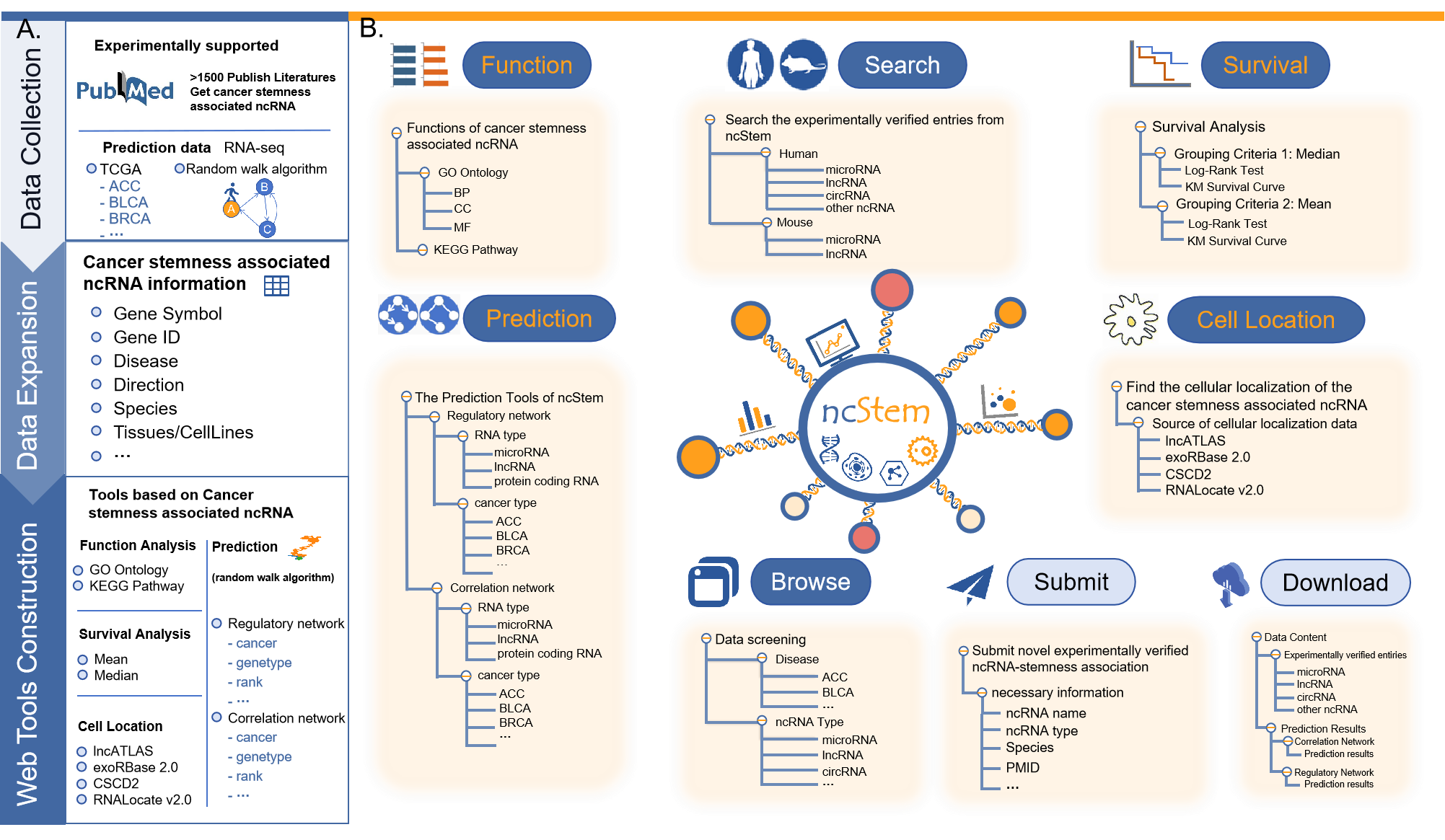
Cancer stemness plays an important role in cancer initiation and progression, and constitutes an important cause of metabolic reprogramming, immune microenvironment remodeling and chemotherapy resistance.
Non-coding RNAs (ncRNAs), including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs), have been proven to be involed in the cancer stemness.
Here, we present ncStem database, which manually curated experimentally validated ncRNAs in cancer stemness.
Users can retrieve detailed information about experimentally verified ncRNAs involved in the cancer stemness, including ncRNA name, ncRNA ID, disease, expression direction and so on.
In addition, computationally predicted cancer stemness associated ncRNAs are also provided in ncStem.
Furthermore, Function, Survival and Cell location tools are integrated in ncStem. Collectively, ncStem offers a user-friendly web server to search and predict ncRNAs in cancer stemness, which might facilitate research on cancer stemness and discover potential targets for cancer treatment.
The home page contains a tool entry, quick search, and brief introduction to help users understand our database.
1. The main functions of the database are provided in the form of menu bars which contains Browse, Search, Tools: Prediction, Function, Survival, Cell Location, Submit, Download, and Help.
2. The brief introduction of ncStem.
3. A quick search for ncRNA information related to cancer stemness was performed by entering an ncRNA name.
4. A summary of the contents of the ncStem database.
5. Data statistics and presentation of ncStem.
6. ncStem provides tools for cancer stemness related ncrnas: prediction tools, enrichment analysis tools, survival analysis tools, and cellular localization tools. Click the button to jump to the corresponding tool page.

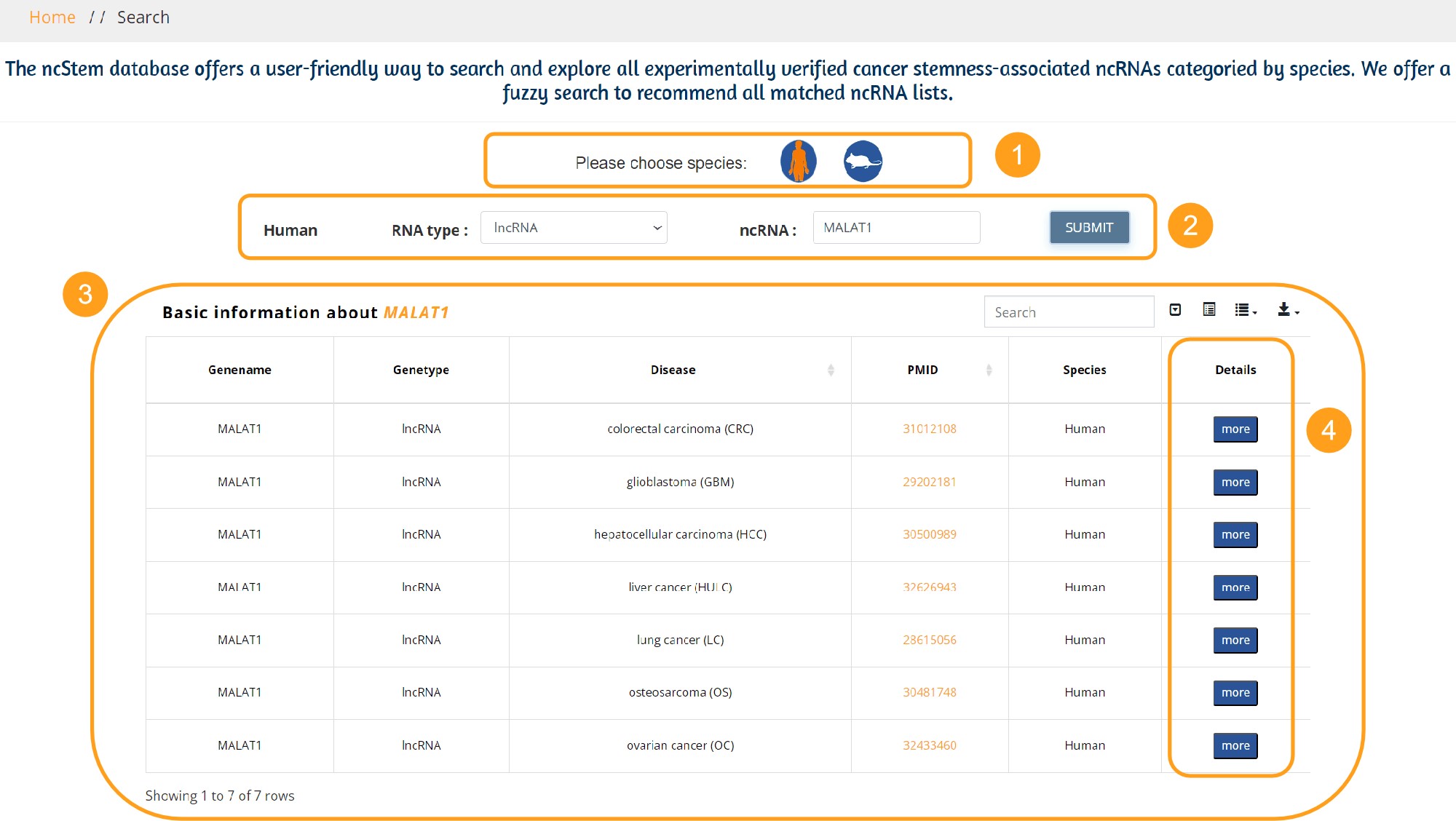
Advanced Search page: Search the experimentally validated entries.
1. ncStem provides a species-based search page, and users can click the icon to select the Human or Mouse search page.
2. Users need to select the ncRNA Type, and the ncRNA list will appear with the corresponding type of experimentally verified cancer stemness associated ncRNA. There are five kinds of ncRNAs (miRNA, lncRNA, circRNA, snoRNA, piRNA). We offer a fuzzy search to recommend all matched miRNA/lncRNA/circRNA/snoRNA/piRNA lists. Clicking the Submit button returns a table with experimentally validated entries.
3. An example of a table with basic entry information is as follows, with genename, genetype, disease, PMID, Species, and a button to jump to with entry details.
4. Users can click the more button to view the table containing the details of the entries, as shown in 5.
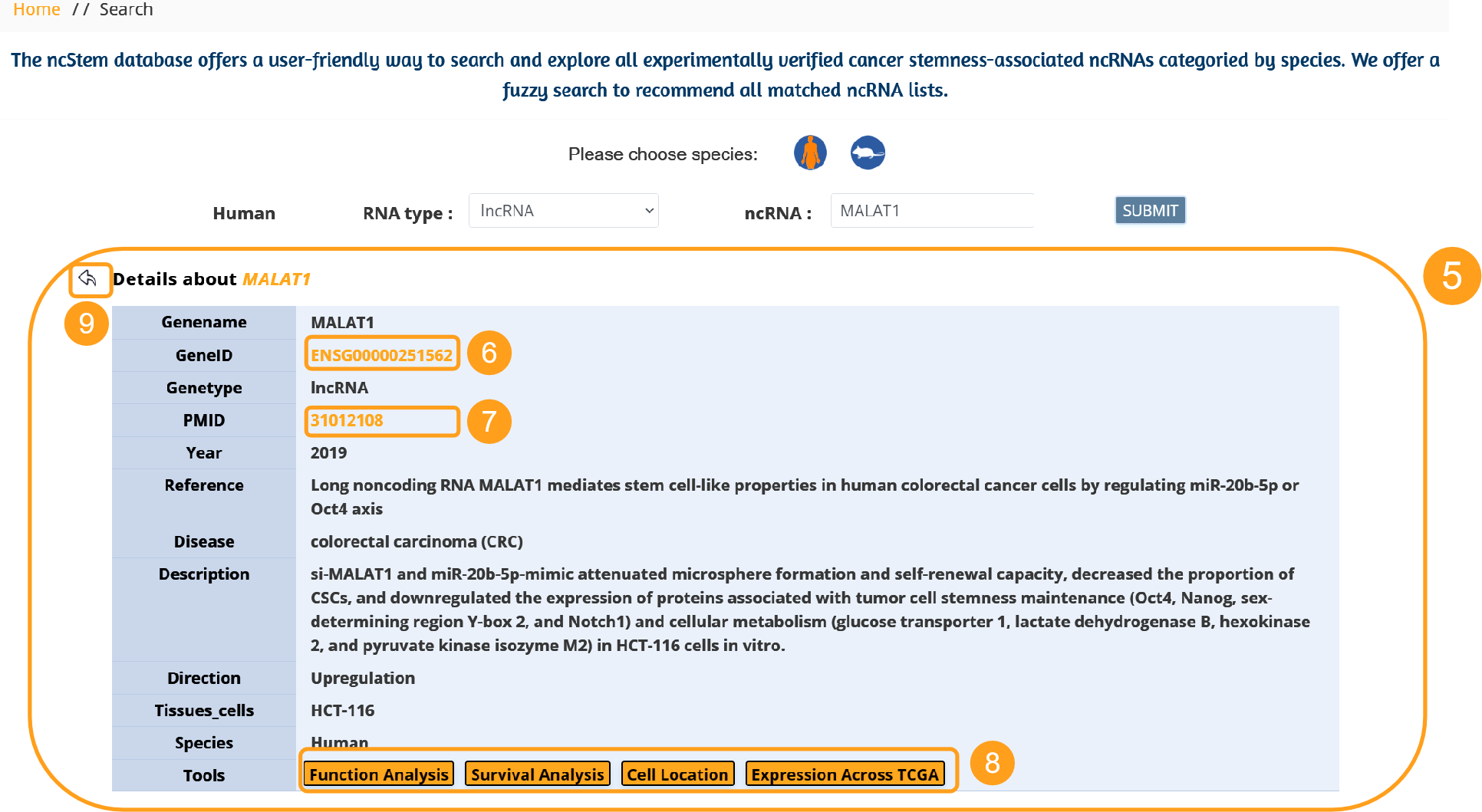
5. Entry details included genename, geneID, genetype, PMID, Year, Reference, Disease, Description, Direction, Tissues/cell lines, Species, Tools.
6. By integrating miRBase, circBase and Ensembl databases, we added corresponding jump links for the ID of cancer stemness associated ncRNA to facilitate users to retrieve more information related to ncRNA.
7. Links are added to the PMID to facilitate users to directly view the literature content.
8. ncStem combined with TCGA database provides Functional Enrichment Analysis, Survival Analysis, and Cellular Localization Analysis tools for experimentally verified cancer stemness associated ncRNAs. Click the button to directly obtain the results of the corresponding analysis tool for the ncRNA.
9. Click on the Back icon to return to the table of basic information on the previous level of entries.
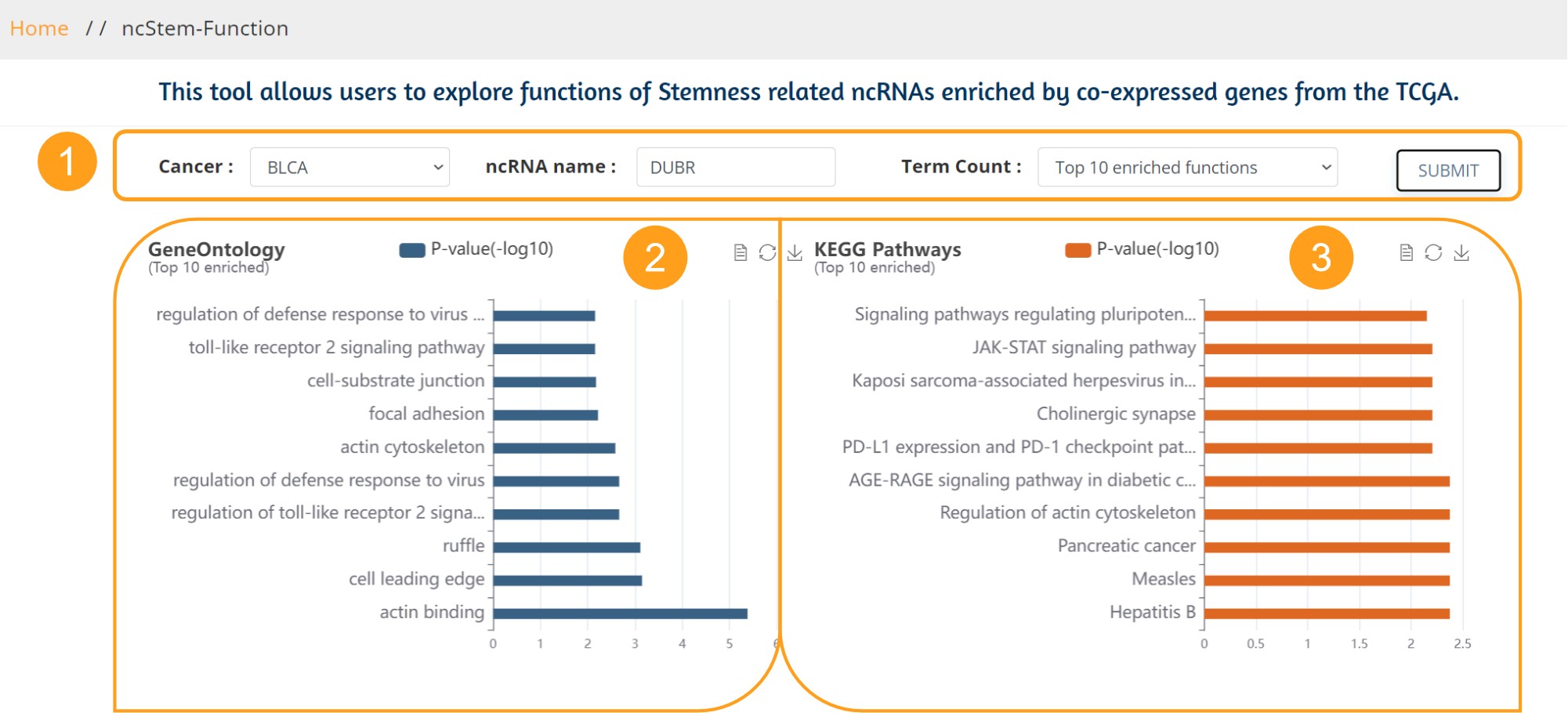
This tool allows users to explore functions of Stemness related ncRNAs enriched by co-expressed genes from the TCGA.
The screening criteria for cancer stemness related ncRNA co-expressed genes were an absolute correlation coefficient greater than 0.5 and a p value less than 0.05.
1. Users can select the cancer of interest from the 33 cancers, and we provide a fuzzy search to recommend a list of all matched ncRNAs, while the users can choose the number of entries to view (Default view top 10). Click the Submit button to return to the analysis results.
2. The enrichment results of Gene Ontology are presented in the form of bar graph, which can be converted to data view, provide download button.
3. The enrichment results of KEGG (Kyoto Encyclopedia of Genes and Genomes)are presented in the form of bar graph, which can be converted to data view, provide download button.
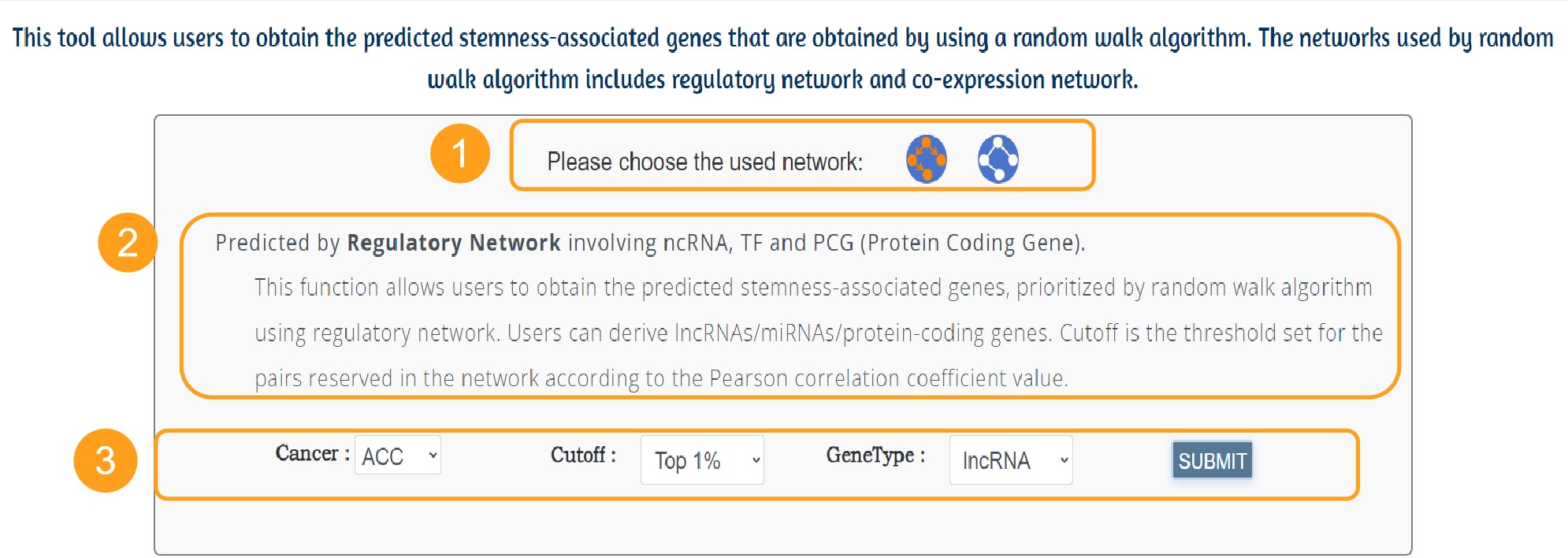
This tool allows users to obtain the predicted stemness-associated genes that are obtained by using a random walk algorithm. The networks used by random walk algorithm includes regulatory network and co-expression network.
1. Users can choose the co-expression network or the regulation network by clicking on the icon.
2. A brief introduction to regulatory or co-expression network prediction tools.
3. Users can select the cancer of interest among the 33 cancers, network cutoffs and the gene type of interest. There are two kinds of ncRNAs (miRNA, lncRNA) to choose from. Click the Submit button to return to the predicted results.
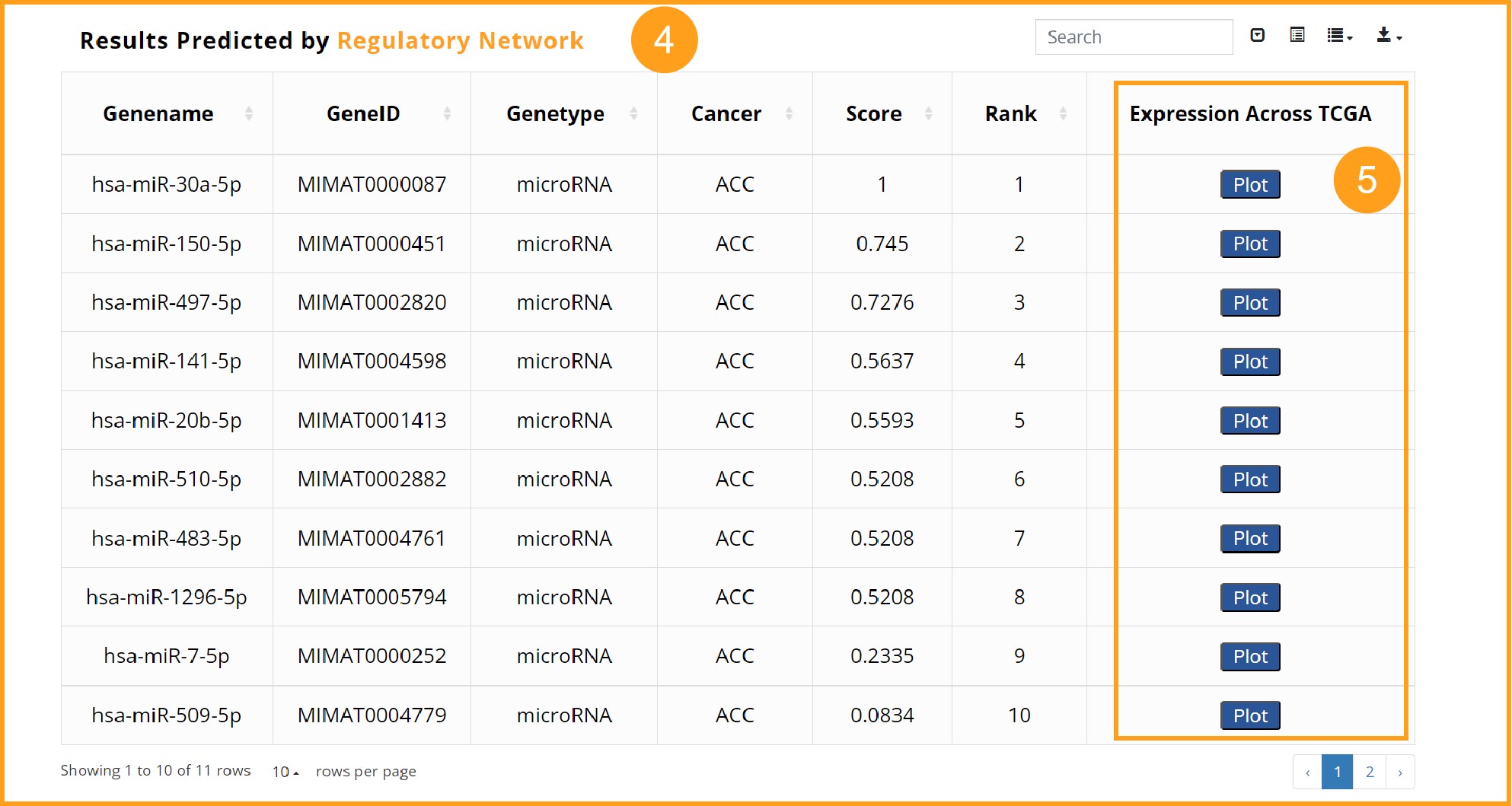
4. An example of the predicted result table. The table, which contains information such as geneID, genename, Genetype, cancer, Score and Rank, is available in a variety of download formats.
5. By clicking the Plot button, the expression of the gene in tumor samples with low stemness index and high stemness index across 33 TCGA cancers can be plotted. If there is a significant difference in expression between the two groups of a cancer, the X-coordinate name will turn red and star.
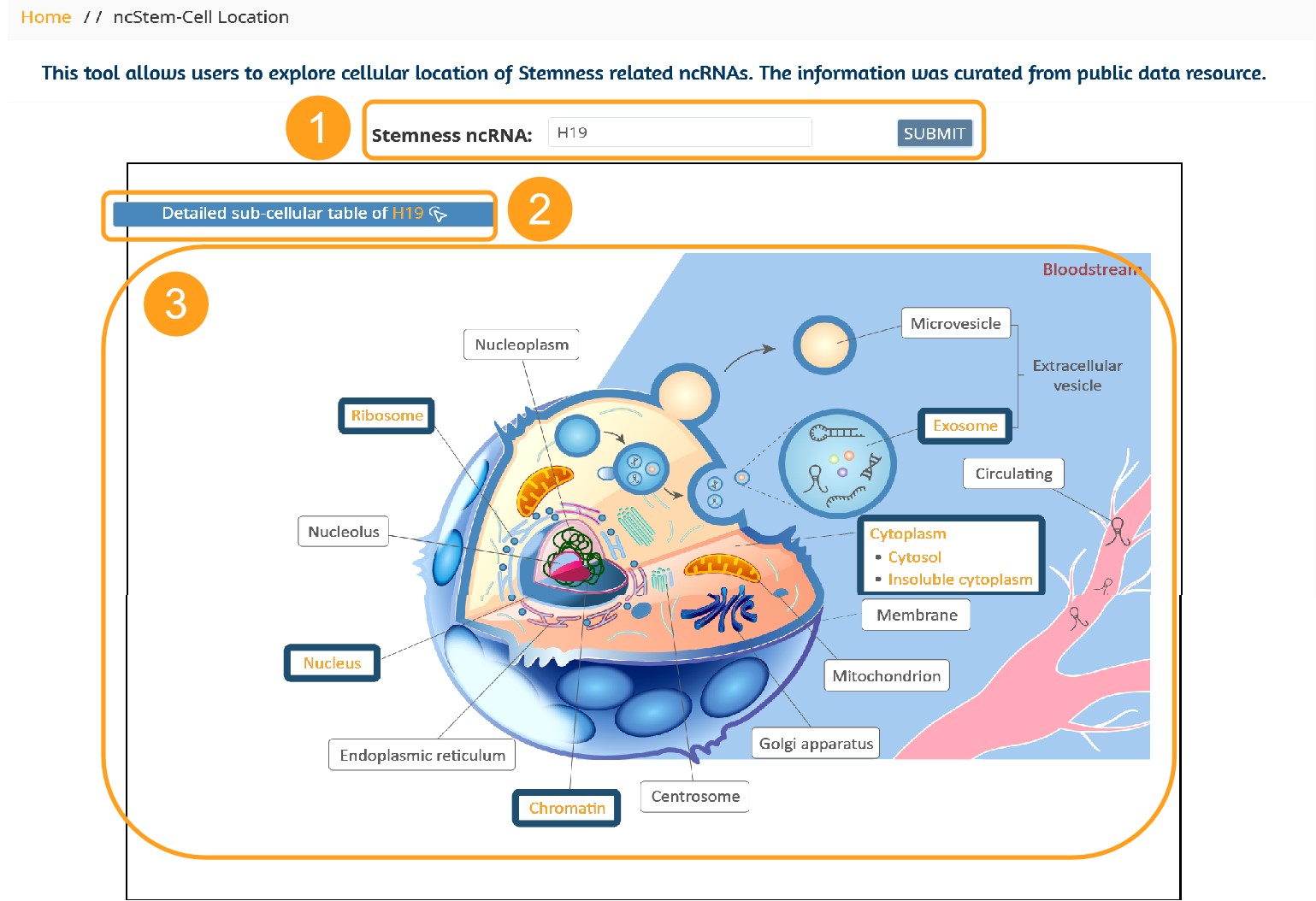
This tool allows users to explore cellular location of Stemness related ncRNAs. The information was curated from public data resource.
1. Users needs to enter the cancer stemness related ncRNA of interest, we provide a fuzzy search to recommend a list of all matched ncRNAs.
2. Click the button to get the floating window to display the cellular localization information of the ncRNA, as shown in 4, including geneID, Location, genename, tissues and source, which can be downloaded in a variety of formats.
3. The corresponding cell position of the ncRNA is given a bold border on the cell vector diagram to provide users with more intuitive cell position information.
4. An example of a suspended window.
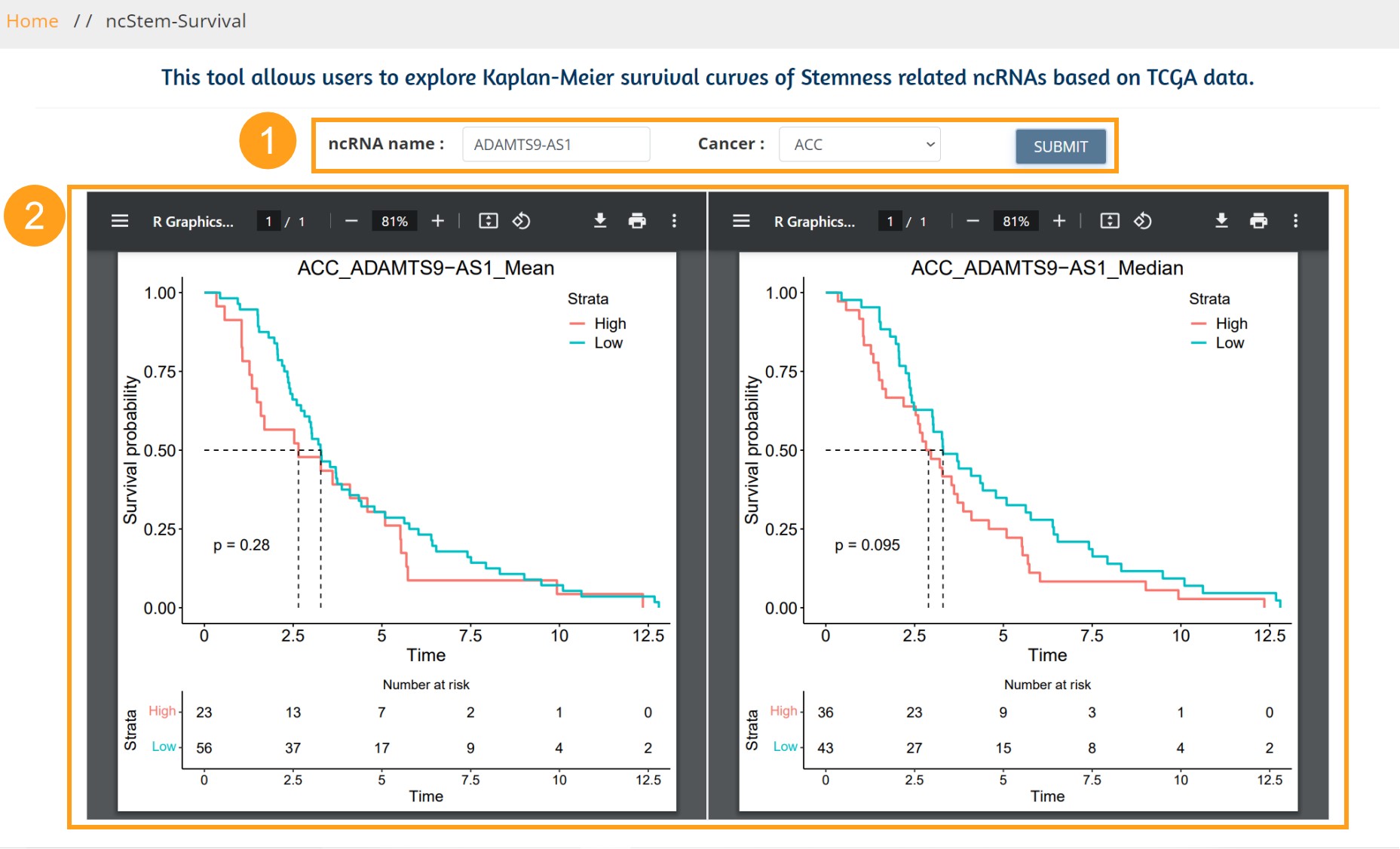
This tool allows users to explore Kaplan-Meier survival curves of Stemness related ncRNAs based on TCGA data.
1. Users need to input the cancer stem-related ncRNA of interest, and we provide fuzzy matching to return the list of ncRNA, and the users need to select the cancer type of interest. Clicking the Submit button starts the online survival plot.
2. ncStem provides survival analysis with two grouping criteria, as shown in the left panel, grouped by the median of the ncRNA in the selected cancer, and the right panel grouped by the mean of the ncRNA in the selected cancer.
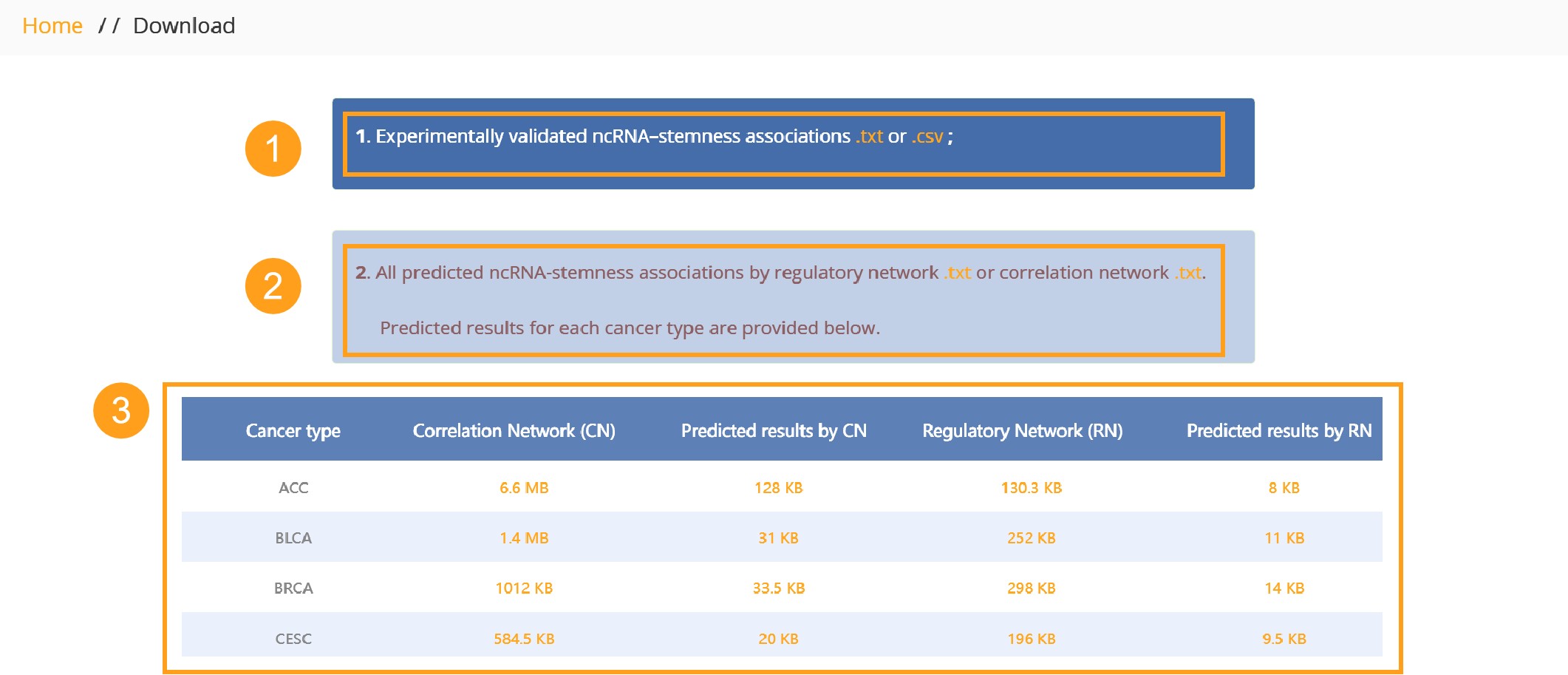
Users could download all of the experimentally verified ncRNA–cancer stemness associations from the ncStem database. In addition, users could download all of the regulation predicted entries, as well as co-expression predicted entries in cancer types.
1. Download all of the experimentally verified ncRNA–cancer stemness associations from the ncStem database.
2. Download all of the regulation predicted entries, as well as co-expression predicted entries in cancer types.
3. Prediction data of cancer stemness related ncRNAs for 33 different cancers: co-expression network data, regulatory network data, results of co-expression network random walk, and results of regulatory network random walk.
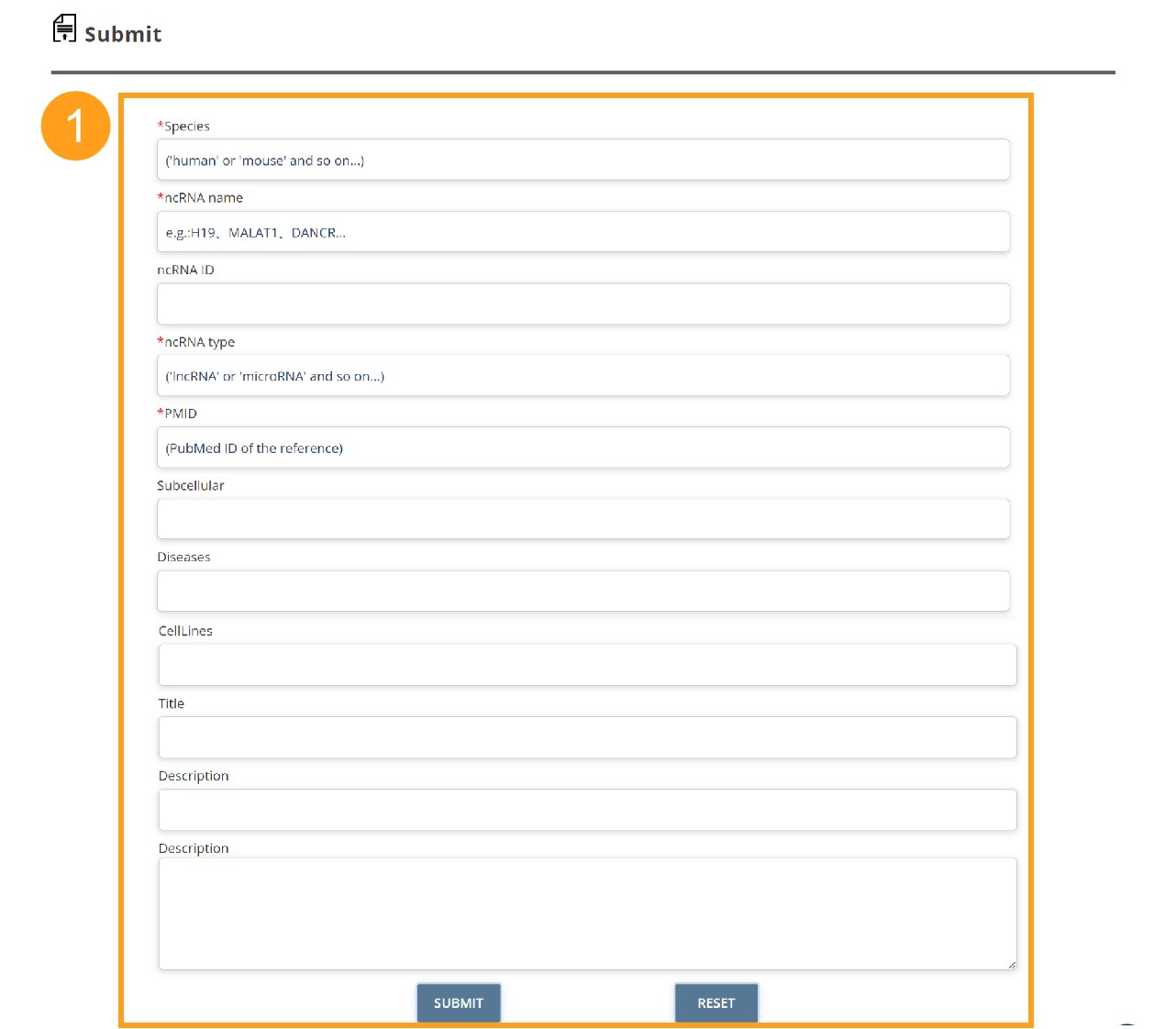
ncStem offers a submission page that enables other researchers to submit experiment verified stemness associated ncRNA data in cancers. Once approved by the submission review committee, the submitted record will be included in the ncStem database and made available to the public in the update release.
1. The information submitted by the user must contain Species, ncRNA name, ncRNA type, and PMID.